ASAF: Optimization of noninvasive assessment of the substrate for atrial fibrillation
ASAF - Optimization of noninvasive assessment of the substrate for atrial fibrillation was a project funded by an International Re-integration Grant from the European Union's Marie-Curie program. The purpose of this project was to use computer modeling to better understand how we can recognize the cardiac conditions that lead to atrial fibrillation. The project was performed in the group of Professor Uli Schotten at Maastricht University.
Introduction
Atrial fibrillation (AF) is a condition in which the atria, the upper chambers of the heart, contract chaotically. AF reduces the pumping efficiency of the heart and makes it beat irregularly. In a healthy heart, an electrical activation mechanism coordinates and tunes the contraction. AF is caused by a malfunction of this activation mechanism. In a normal heart the activation is generated at regular intervals in a single location, called the sinus node, and from there spreads over the atria. In AF, the activation can run in circles, resulting in a chaotic and irregular contraction. AF maintains and aggravates itself by causing electrophysiological and structural changes. Treatment options depend on the disease stage. Our purpose is to develop better diagnostic methods to identify the stage of AF in individual patients.
The structural remodeling process that maintains AF causes heterogeneous changes in the atrial tissue's capacity to propagate the electrical activation. Therefore, the complexity of the propagating activation wavefront could be a measure of the disease stage. It would be desirable to judge this complexity from the ECG measured at the body surface. We hypothesized that more complex conduction patterns in the atria generate more complex potential patterns on the body surface. The purpose of our project was to find and validate measures for the complexity of ECG signals that correlate well with the complexity of wavefront propagation in the atria. To achieve this, we developed a large-scale computer model of the human atria with which we could accurately simulate the atrial activation pattern and the electrical signals (ECG and local electrograms) that result from it.
Results of this project
We have built an anatomical model of the atria that represents the structural inhomogeneities that are present even in the healthy atria, and that allows pathological changes in the structure to be represented. The model includes a clear representation of the thin atrial wall and the thicker muscle bundles that are invariably present. With this model we simulated the electrical activation mechanism of the atria based on the transmembrane ionic currents in the atrial muscle cells. From the results we computed the ECG together with electrograms in the atria (which can be measured with endocardial catheters in real patients) as well as electrograms in the esophagus (which can also be measured in patients using a special probe).
We discovered that the model could not reproduce the atrial ECG as it is commonly described in cardiology textbooks and observed in clinical ECGs. Even after a long period of tuning, the simulated ECG remained considerably more complex than what is seen on the clinical ECG. However, we were aware that clinical ECGs are electronically filtered to remove noise and interference - unwanted signals that are almost as large as the atrial ECG itself. We suspected that these filters hide much of the complexity that is present in the real physiological signals. We therefore decided to measure ECGs ourselves, using techniques that avoid filtering. We used specialised high-quality ECG equipment to record ECGs with extremely low noise and interference levels. Moreover, we made long recordings (5 to 10 minutes) to obtain hundreds of heartbeats. These recordings were cut into single-beat segments, each aligned on the P wave (the part of the ECG generated by the atria). In each of these segments, the physiological signal is supposed to be the same, while the noise and interference are different each time. Therefore, when the average of hundreds of segments is computed, the real signals add up while the noise and interference cancel out. Thus a very clean signal is obtained without any filtering. We performed this analysis on normal volunteers of different ages.
The P waves that we obtained were very different from the filtered signals that are seen in the clinic. Despite the absence of heart disease, the signals were very complex, with 2 to 10 peaks appearing in a single P wave, even in the youngest subjects.
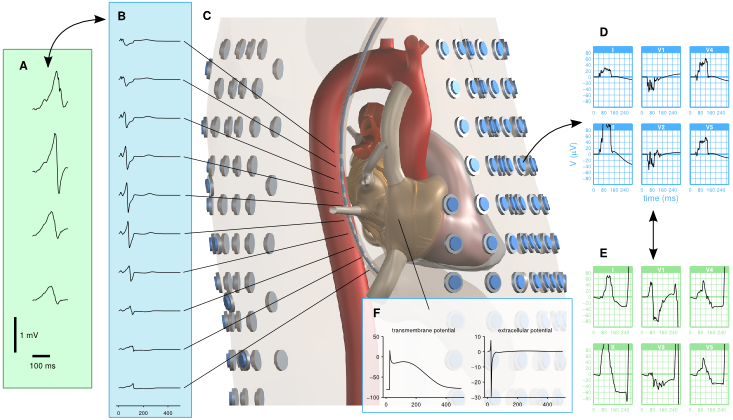
The figure above illustrates how measured trans-esophagal ECGs (panel A) are compared with their simulated equivalents (B) extracted from a virtual probe in the torso model (C). Surface ECGs are extracted from hundreds of surface locations (D) and compared to measured ECGs obtained with the same electrode set in human volunteers (E). The model also computes several local signals such as transmembrane potentials and local electrograms (F), which can be compared to data from other studies.
Fibrillation studies
Now that we know that our model reproduces the normal atrial ECG well, we have started the simulations of AF that we originally intended to perform. The movie below shows an example, performed with an intermediate version of the model.
We run many simulations of this type, with different degrees of tissue damage, to learn how this affects the complexity of the activation pattern and the surface ECG.