Heart Model: features and statistics
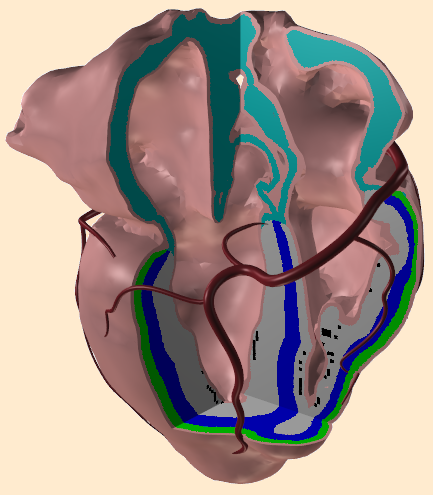
Some features of our model are
- Anatomy obtained from CT scans as described by Lorange and Gulrajani in 1993, but ...
- we are presently using a refined finite-difference grid with 0.2-mm resolution...
- which results in 25 million to 60 million nodes...
- consuming 12 GB of computer memory for a monodomain, and 60 GB for a bidomain simulation.
- The Purkinje system is modeled using early activation times obtained from the human heart studies of Durrer et al 1970
- Several membrane models are interfaced to our model, such as those by Bernus and by Ten Tusscher.
- The bidomain equations are solved using a fully-explicit integration of the membrane potential, and a BiCGSTAB algorithm with ILU(0) preconditioning is used to solve for the extracellular potential
- All this is currently done on 64 processors of an SGI Altix 4700 computer at RQCHP, which does a monodomain simulation of a single heart beat in 40 minutes and a bidomain simulation in about 1 day.
- Our monodomain model is described in detail by Trudel et al. IEEE Trans Biomed Eng 2004; recent improvements in Potse, Dubé and Gulrajani, Conf IEEE-EMBS 2003
- We first reported on our bidomain model at the 31st International Congress on Electrocardiology June/July 2004 in Kyoto, Japan. A more substantial paper about this model was published in IEEE Trans Biomed Eng 2006. For more recent publications see my list of papers.